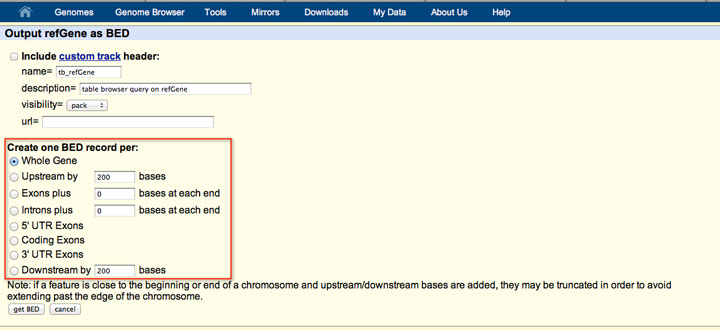
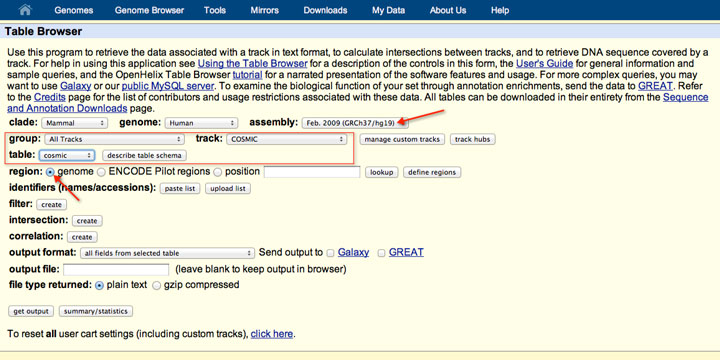
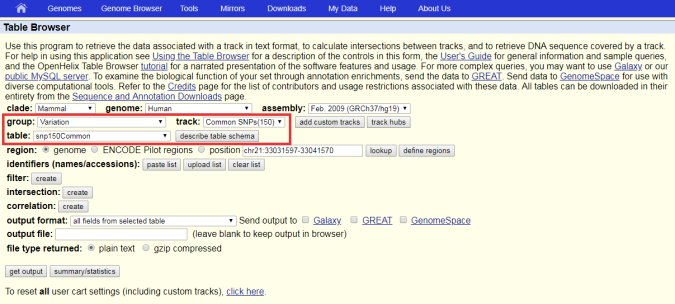
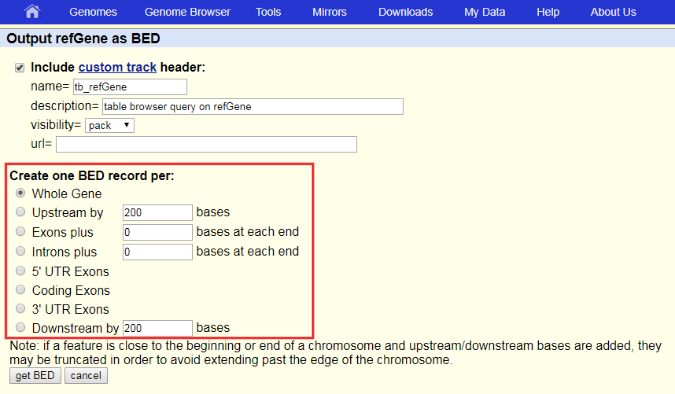
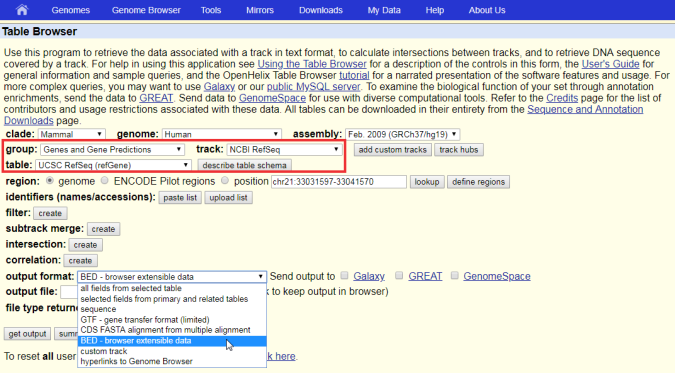
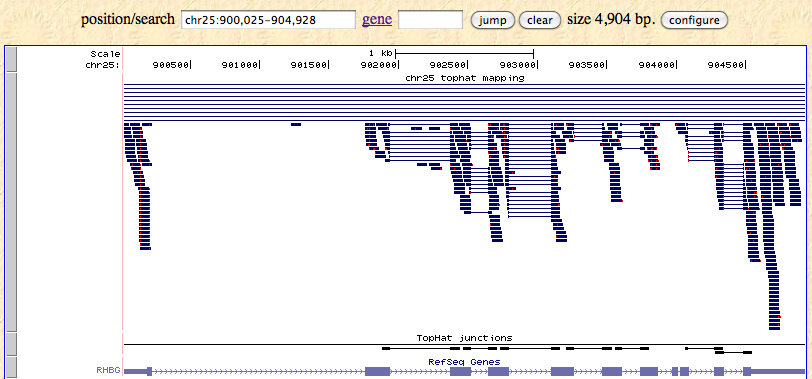
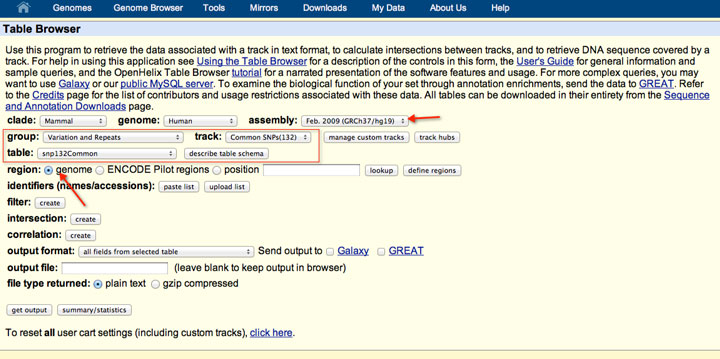
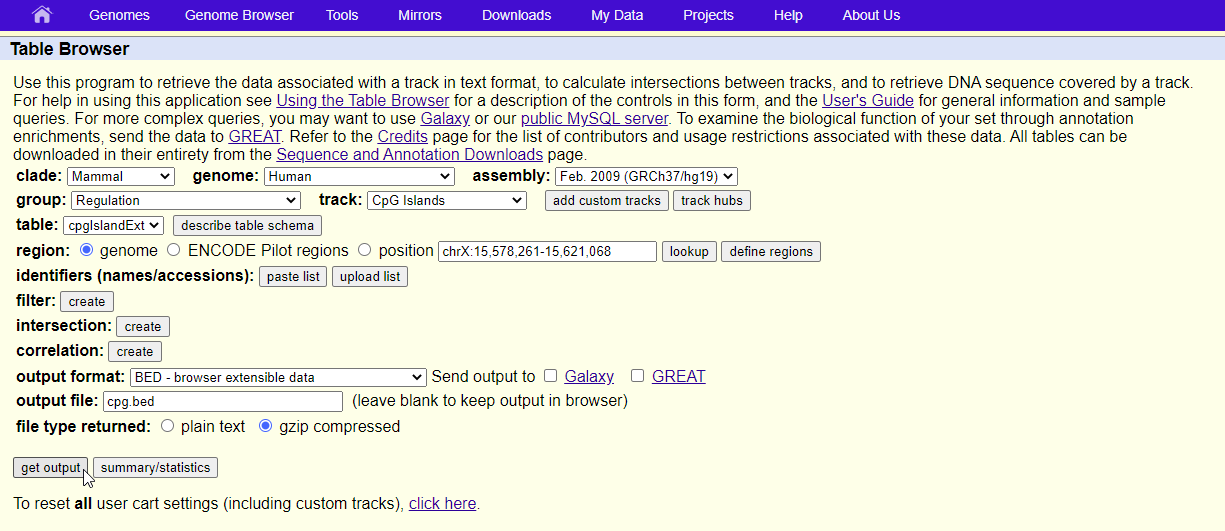
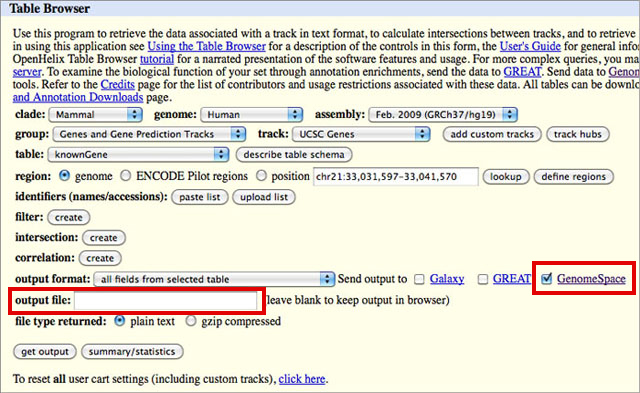
How to download genomic coordinates for transposons/RepeatMasker from UCSC genome browser - Min Dai - Medium

The output of the alignment procedure is stored in a text file [BED... | Download Scientific Diagram

Bedgraph visualization using the UCSC genome browser of the PITX1 gene... | Download Scientific Diagram